Schema.org, BioSchemas, JSONSchema, JSON-LD and DATS
Contents
Schema.org, BioSchemas, JSONSchema, JSON-LD and DATS¶
A case study with KidsFirst is considered for asset metadata serialization as DATS, and validation with JSONSchema
Authors: Daniel J. B. Clarke
Maintainers: Daniel J. B. Clarke
Version: 1.0
License: GPLv2+
Motivations¶
The Data Tag Suite (DATS) metadata model as described in this paper and fully codefied in this repository strives to model datasets irrespective of their domains. DATS embodies several key elements making it extroudinarily useful for FAIRification:
Machine Readibility: Datasets described with a consistent DATS format permit machines to resolve FAIR metadata such as identifiers, authorship, funding, citation, license, consent, access, provenance, and ultimatly topic as well.
RDF Interoperability: Serialized in strict JSON-LD, the DATS format is renderable as an RDF graph permitting interoperability with ontological vocabularies and existing dataset description formats including schema.org and the Open Biological and Biomedical Ontology (OBO).
Findability: The utilization of these consistent formats will permit various existing services and endless future ones to be able to identify aspects of the dataset for the purposes of indexing and searching. One such service is google dataset search which utilizes schema.org metadata.
CFDE Compatibility: Tooling has been created to convert DATS to the C2M2 and for automatically evaluating the FAIRness of Datasets through the DATS metadata model.
Objectives¶
Preparation¶
We need to get the manifest or access to an API serving the existing
metadata. In our case study, KidsFirst,
the assets are browsable in the file repository.
After enabling all “Columns” click the “Export TSV” button and save that file to
./data/file-table.tsv.
# Python tool for data table processing
import pandas as pd
# Jupyter Notebook display helper
from IPython.display import display
df = pd.read_csv('./data/file-table.tsv', sep='\t', low_memory=False)
display(df.head())
| File ID | Participants ID | Study Name | Proband | Family Id | Data Type | File Format | File Size | Participant External ID | File Name | File External ID | Aliquot External ID | Sample External ID | Biospecimen ID | Tissue Type (Source Text) | Diagnosis (Source Text) | Study ID | Latest DID | Observed | Repository | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | GF_000VDK42 | PT_PR4YBBH3 | Pediatric Brain Tumor Atlas: CBTTC | Yes | -- | Aligned Reads | bam | 11041645308 | C29274 | 62c0c6fe-99f8-4ff7-b3b5-233e6cc2ff0f.bam | 62c0c6fe-99f8-4ff7-b3b5-233e6cc2ff0f.bam | 746063 | 7316-126-T-112502.RNA-Seq | BS_A7Q8G0Y1 | Tumor | Brainstem glioma- Diffuse intrinsic pontine gl... | SD_BHJXBDQK | bdf6c2f6-1500-4693-ae32-fd18dc4ab9e1 | -- | gen3 |
| 1 | GF_000WBJCD | PT_NK8A49X5 | Pediatric Brain Tumor Atlas: PNOC | Yes | -- | Annotated Somatic Mutations | maf | 122552 | P-06 | 77af1324-3754-4e34-a208-d1342a2f2ca6.mutect2_s... | harmonized/simple-variants/77af1324-3754-4e34-... | A08713, A08710 | A08692-T.WXS, A08691-N.WXS | BS_6DT506HY, BS_5DPMQQVG | Tumor, Normal | Brainstem glioma- Diffuse intrinsic pontine gl... | SD_M3DBXD12 | 16712090-50f7-4cd1-bf2d-90ce989c2139 | -- | gen3 |
| 2 | GF_001JWT9N | PT_G16VK7FR | Pediatric Brain Tumor Atlas: PNOC | Yes | -- | Gene Fusions | 5779507 | P-37 | 9f586dc1-1df8-4f59-9a01-803141fffb94.arriba.fu... | harmonized/gene-fusions/9f586dc1-1df8-4f59-9a0... | A19683 | A19649-T.RNA-Seq | BS_XGDPK33A | Tumor | Brainstem glioma- Diffuse intrinsic pontine gl... | SD_M3DBXD12 | 49f7aead-ce23-4c38-b566-1d99cb5a5435 | -- | gen3 | |
| 3 | GF_002DRSGP | PT_2HN13G42 | Kids First: Congenital Diaphragmatic Hernia | Yes | FM_F9S808PW | Aligned Reads | cram | 23537329440 | CDH14-0006 | CDH14-0006.cram | s3://kf-study-us-east-1-prd-sd-46sk55a3/source... | CDH14-0006 | CDH14-0006 | BS_BKH9S8YN | Normal | congential diaphragmatic hernia | SD_46SK55A3 | c4c9d542-21fb-487d-ac07-916d466774a8 | true, false, false, false | gen3 |
| 4 | GF_004J173A | PT_MH56TZJD | Kids First: Congenital Diaphragmatic Hernia | No | FM_1EFM6M40 | Aligned Reads | cram | 17290282717 | CDH4-84F | CDH4-84F.cram | s3://kf-study-us-east-1-prd-sd-46sk55a3/source... | CDH4-84F | CDH4-84F | BS_MP5P3ZPH | Normal | -- | SD_46SK55A3 | 02642bc8-ae46-45a1-8932-2765cf1df480 | -- | gen3 |
DATS Conversion¶
The full DATS schema is available here, it includes a JSON Schema definition as well as a visualization of how things fit together.

DATS uses a strict JSON-LD serialization.
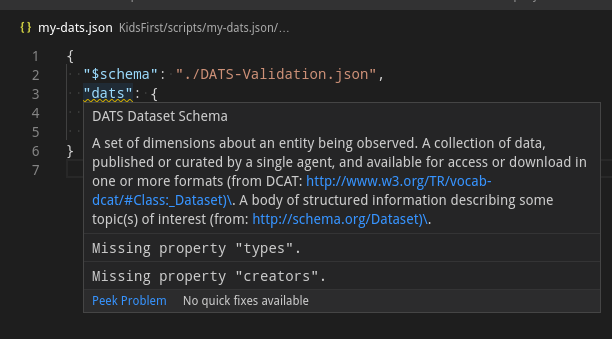
There are several ways to get a sense of what the metadata model entails. In some cases starting with an example is easier, but everything is easier with autocompletion and type-hints. Several code editors support JSON Schema for auto completion (see this).
With visual studio code, you can set this up by linking to the
schema with a $schema field.
%%sh
# Create a file DATS-Validation.json with the following contents
# this is a simple json-schema which references the public DATS schema validator
cat > DATS-Validation.json << EOF
{
"\$schema": "http://json-schema.org/draft-04/schema",
"type": "object",
"properties": {
"dats": {
"\$ref": "https://raw.githubusercontent.com/datatagsuite/schema/master/dataset_schema.json"
}
}
}
EOF
# Create a file to edit which will validate against the file from the DATS-Validation file
cat > my-dats.json << EOF
{
"\$schema": "./DATS-Validation.json",
"dats": {
"title": "My First Json-Schema Validated DATS Object"
}
}
EOF
Modifying the created my-dats.json, you should be able to explore the fields
through autocompletion with an editor that supports it.
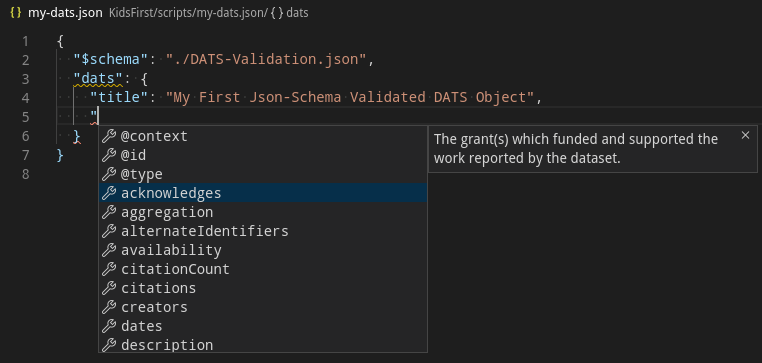
Another way, or perhaps also, is to learn by example. Several other DCC’s assets were
processed and converted to DATS here, example files
and scripts can be found in the DCC_name/output and DCC_name/scripts directories
respectively.
It’s now time to convert what we can into DATS, striving to capture as much as possible from the original table.
Challenge 1: What do you mean by Dataset?¶
Even in this case, the definition of a Dataset becomes problematic and unclear. Remember that things we codify are often models and as such are not always perfect. Rather than thinking about Dataset with your interpretation of what it is, think of it in terms of how it will end up being used.
This is what a ‘dataset’ looks like on Google Dataset Search; the same fields will be used, and more; for your own assets.
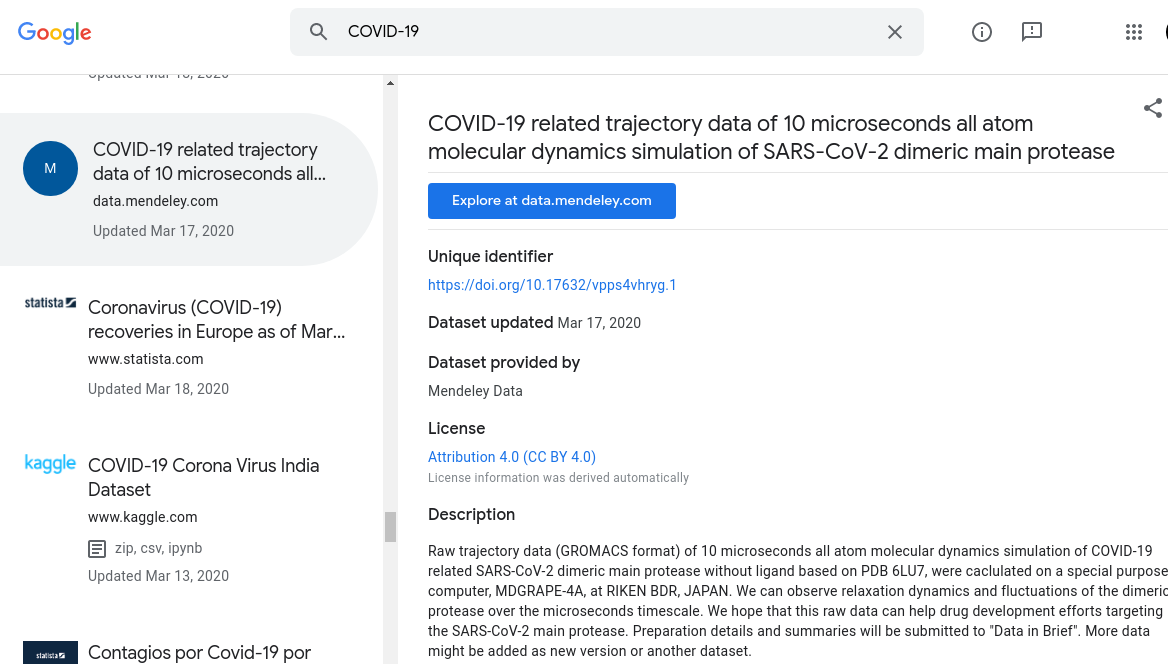
In other words, irrespective of what your definition is of a ‘dataset’, you should consider using something that is identifiable enough to have its own unique metadata including dedicated landing page, unique identifier, citation, license, and more. File assets associated with that dataset will be listed under the dataset.
Importantly, Datasets should ideally be associatable with singular biosamples when possible, so in some cases, it may make sense to consider each individual file to be its own dataset if each individual file is actually established for every biosample.
Do note that DATS also supports Dataset in Dataset relationships if that becomes necessary.
# The KidsFirst table has 5 primary entity types in this file and a unique identifier
display(df[['File ID', 'Participants ID', 'Study ID', 'Biospecimen ID', 'Latest DID']].head())
# Using JSON-LD and keeping in mind the arbitrary DATS structure,
# things should end up looking like so:
def dats_from_record(record):
return {
'@type': 'Dataset',
'identifier': {
'@type': 'Identifier',
'identifier': record['Latest DID'],
},
'producedBy': {
# The dataset in question was produced as part of a study
'@type': 'Study',
'identifier': {
'@type': 'Identifier',
'identifier': record['Study ID'],
},
},
'isAbout': [
{
# The dataset in question has a biospecimen
'@type': 'BiologicalEntity',
'identifier': {
'@type': 'Identifier',
'identifier': record['Biospecimen ID'],
},
},
{
# The dataset in question is about this participant
'@type': 'StudyGroup',
'identifier': {
'@type': 'Identifier',
'identifier': record['Participants ID'],
},
},
],
'distributions': [
{
# The dataset in question has this file
'@type': 'DatasetDistribution',
'identifier': {
'@type': 'Identifier',
'identifier': record['File ID'],
},
}
],
}
# Converting each element to DATS
dats = {
# schema.org context, gives RDF meaning to `@type` and `predicates` as defined by schema.org
'@context': 'http://w3id.org/dats/context/sdo/dataset_sdo_context.jsonld',
'@graph': [
dats_from_record(record)
for _, record in df.head().iterrows()
]
}
display(dats)
| File ID | Participants ID | Study ID | Biospecimen ID | Latest DID | |
|---|---|---|---|---|---|
| 0 | GF_000VDK42 | PT_PR4YBBH3 | SD_BHJXBDQK | BS_A7Q8G0Y1 | bdf6c2f6-1500-4693-ae32-fd18dc4ab9e1 |
| 1 | GF_000WBJCD | PT_NK8A49X5 | SD_M3DBXD12 | BS_6DT506HY, BS_5DPMQQVG | 16712090-50f7-4cd1-bf2d-90ce989c2139 |
| 2 | GF_001JWT9N | PT_G16VK7FR | SD_M3DBXD12 | BS_XGDPK33A | 49f7aead-ce23-4c38-b566-1d99cb5a5435 |
| 3 | GF_002DRSGP | PT_2HN13G42 | SD_46SK55A3 | BS_BKH9S8YN | c4c9d542-21fb-487d-ac07-916d466774a8 |
| 4 | GF_004J173A | PT_MH56TZJD | SD_46SK55A3 | BS_MP5P3ZPH | 02642bc8-ae46-45a1-8932-2765cf1df480 |
{'@context': 'http://w3id.org/dats/context/sdo/dataset_sdo_context.jsonld',
'@graph': [{'@type': 'Dataset',
'identifier': {'@type': 'Identifier',
'identifier': 'bdf6c2f6-1500-4693-ae32-fd18dc4ab9e1'},
'producedBy': {'@type': 'Study',
'identifier': {'@type': 'Identifier', 'identifier': 'SD_BHJXBDQK'}},
'isAbout': [{'@type': 'BiologicalEntity',
'identifier': {'@type': 'Identifier', 'identifier': 'BS_A7Q8G0Y1'}},
{'@type': 'StudyGroup',
'identifier': {'@type': 'Identifier', 'identifier': 'PT_PR4YBBH3'}}],
'distributions': [{'@type': 'DatasetDistribution',
'identifier': {'@type': 'Identifier', 'identifier': 'GF_000VDK42'}}]},
{'@type': 'Dataset',
'identifier': {'@type': 'Identifier',
'identifier': '16712090-50f7-4cd1-bf2d-90ce989c2139'},
'producedBy': {'@type': 'Study',
'identifier': {'@type': 'Identifier', 'identifier': 'SD_M3DBXD12'}},
'isAbout': [{'@type': 'BiologicalEntity',
'identifier': {'@type': 'Identifier',
'identifier': 'BS_6DT506HY, BS_5DPMQQVG'}},
{'@type': 'StudyGroup',
'identifier': {'@type': 'Identifier', 'identifier': 'PT_NK8A49X5'}}],
'distributions': [{'@type': 'DatasetDistribution',
'identifier': {'@type': 'Identifier', 'identifier': 'GF_000WBJCD'}}]},
{'@type': 'Dataset',
'identifier': {'@type': 'Identifier',
'identifier': '49f7aead-ce23-4c38-b566-1d99cb5a5435'},
'producedBy': {'@type': 'Study',
'identifier': {'@type': 'Identifier', 'identifier': 'SD_M3DBXD12'}},
'isAbout': [{'@type': 'BiologicalEntity',
'identifier': {'@type': 'Identifier', 'identifier': 'BS_XGDPK33A'}},
{'@type': 'StudyGroup',
'identifier': {'@type': 'Identifier', 'identifier': 'PT_G16VK7FR'}}],
'distributions': [{'@type': 'DatasetDistribution',
'identifier': {'@type': 'Identifier', 'identifier': 'GF_001JWT9N'}}]},
{'@type': 'Dataset',
'identifier': {'@type': 'Identifier',
'identifier': 'c4c9d542-21fb-487d-ac07-916d466774a8'},
'producedBy': {'@type': 'Study',
'identifier': {'@type': 'Identifier', 'identifier': 'SD_46SK55A3'}},
'isAbout': [{'@type': 'BiologicalEntity',
'identifier': {'@type': 'Identifier', 'identifier': 'BS_BKH9S8YN'}},
{'@type': 'StudyGroup',
'identifier': {'@type': 'Identifier', 'identifier': 'PT_2HN13G42'}}],
'distributions': [{'@type': 'DatasetDistribution',
'identifier': {'@type': 'Identifier', 'identifier': 'GF_002DRSGP'}}]},
{'@type': 'Dataset',
'identifier': {'@type': 'Identifier',
'identifier': '02642bc8-ae46-45a1-8932-2765cf1df480'},
'producedBy': {'@type': 'Study',
'identifier': {'@type': 'Identifier', 'identifier': 'SD_46SK55A3'}},
'isAbout': [{'@type': 'BiologicalEntity',
'identifier': {'@type': 'Identifier', 'identifier': 'BS_MP5P3ZPH'}},
{'@type': 'StudyGroup',
'identifier': {'@type': 'Identifier', 'identifier': 'PT_MH56TZJD'}}],
'distributions': [{'@type': 'DatasetDistribution',
'identifier': {'@type': 'Identifier', 'identifier': 'GF_004J173A'}}]}]}
There are several improvements we can make to the above:
Give context to our identifiers, which only make sense in the context of KidsFirst
Provide more metadata as available in our table
def dats_from_record(record):
return {
'@type': 'Dataset',
'identifier': {
'@type': 'Identifier',
'identifier': record['Latest DID'],
'identifierSource': 'https://portal.kidsfirstdrc.org/',
},
'storedIn': {
'@type': 'DataRepository',
'name': record['Repository'],
},
'producedBy': {
'@type': 'Study',
'identifier': {
'@type': 'Identifier',
'identifier': record['Study ID'],
'identifierSource': 'https://portal.kidsfirstdrc.org/',
},
'name': record['Study Name'],
},
'isAbout': [
{
'@type': 'BiologicalEntity',
'identifier': {
'@type': 'Identifier',
'identifier': record['Biospecimen ID'],
'identifierSource': 'https://portal.kidsfirstdrc.org/',
},
'alternateIdentifiers': [
{
'@type': 'AlternateIdentifier',
'identifier': record['Sample External ID'],
# NOTE: Preferred identifierSource with globally unique semantic URI
},
{
'@type': 'AlternateIdentifier',
'identifier': record['Aliquot External ID'],
# NOTE: Preferred identifierSource with globally unique semantic URI
},
],
},
{
'@type': 'StudyGroup',
'identifier': {
# NOTE: Ideally, `${identifierSource}${identifier}` resolves to a landing page for this entity
'@type': 'Identifier',
'identifier': record['Participants ID'],
'identifierSource': 'https://portal.kidsfirstdrc.org/participant/',
},
'alternateIdentifiers': [
{
'@type': 'AlternateIdentifier',
'identifier': record['Participant External ID'],
# NOTE: Preferred identifierSource with globally unique semantic URI
},
],
},
],
'distributions': [
{
'identifier': {
# NOTE: Ideally, `${identifierSource}${identifier}` resolves to a landing page for this entity
'@type': 'Identifier',
'identifier': record['File ID'],
'identifierSource': 'https://portal.kidsfirstdrc.org/file/',
},
'@type': 'DatasetDistribution',
'formats': [
record['File Format'],
],
'size': record['File Size'],
'unit': {
'@type': 'Annotation',
'value': 'bytes',
# NOTE: Preferred valueIRI with globally unique semantic URI
},
'access': {
'@type': 'Access',
'identifier': {
'@type': 'Identifier',
'identifier': record['File Name'],
},
'alternateIdentifiers': [
{
'@type': 'AlternateIdentifier',
'identifier': record['File External ID'],
# NOTE: Preferred identifierSource with globally unique semantic URI
},
],
'landingPage': 'https://portal.kidsfirstdrc.org/file/' + record['File ID'],
# NOTE: Ideally accessURL would be specified
}
}
],
'types': [
{
'@type': 'DataType',
'information': {
'@type': 'Annotation',
'value': record['Data Type'],
},
},
],
'extraProperties': [*filter(None, [
# Metadata that doesn't fit anywhere else in DATS but may be relevant
{
'@type': 'CategoryValuesPair',
'category': 'tissue',
# NOTE: Preferred categoryIRI with globally unique semantic URI
'values': [
{
'@type': 'Annotation',
'value': record['Tissue Type (Source Text)'],
# NOTE: Preferred valueIRI with globally unique semantic URI
}
]
} if record['Tissue Type (Source Text)'] != '--' else None,
{
'@type': 'CategoryValuesPair',
'category': 'diagnosis',
# NOTE: Preferred categoryIRI with globally unique semantic URI
'values': [
{
'@type': 'Annotation',
'value': record['Diagnosis (Source Text)'],
# NOTE: Preferred valueIRI with globally unique semantic URI
}
]
} if record['Diagnosis (Source Text)'] != '--' else None, # Don't create entries for junk
{
'@type': 'CategoryValuesPair',
'category': 'proband',
# NOTE: Preferred categoryIRI with globally unique semantic URI
'values': [
{
'@type': 'Annotation',
'value': record['Proband'],
# NOTE: Preferred valueIRI with globally unique semantic URI
},
],
} if record['Proband'] != '--' else None, # Don't create entries for junk,
])],
}
# Converting each element to DATS
dats = {
# schema.org context, gives RDF meaning to `@type` and `predicates` as defined by schema.org
'@context': 'http://w3id.org/dats/context/sdo/dataset_sdo_context.jsonld',
'@graph': [
dats_from_record(record)
for _, record in df.head().iterrows()
]
}
display(dats)
{'@context': 'http://w3id.org/dats/context/sdo/dataset_sdo_context.jsonld',
'@graph': [{'@type': 'Dataset',
'identifier': {'@type': 'Identifier',
'identifier': 'bdf6c2f6-1500-4693-ae32-fd18dc4ab9e1',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'storedIn': {'@type': 'DataRepository', 'name': 'gen3'},
'producedBy': {'@type': 'Study',
'identifier': {'@type': 'Identifier',
'identifier': 'SD_BHJXBDQK',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'name': 'Pediatric Brain Tumor Atlas: CBTTC'},
'isAbout': [{'@type': 'BiologicalEntity',
'identifier': {'@type': 'Identifier',
'identifier': 'BS_A7Q8G0Y1',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': '7316-126-T-112502.RNA-Seq'},
{'@type': 'AlternateIdentifier', 'identifier': '746063'}]},
{'@type': 'StudyGroup',
'identifier': {'@type': 'Identifier',
'identifier': 'PT_PR4YBBH3',
'identifierSource': 'https://portal.kidsfirstdrc.org/participant/'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'C29274'}]}],
'distributions': [{'identifier': {'@type': 'Identifier',
'identifier': 'GF_000VDK42',
'identifierSource': 'https://portal.kidsfirstdrc.org/file/'},
'@type': 'DatasetDistribution',
'formats': ['bam'],
'size': 11041645308,
'unit': {'@type': 'Annotation', 'value': 'bytes'},
'access': {'@type': 'Access',
'identifier': {'@type': 'Identifier',
'identifier': '62c0c6fe-99f8-4ff7-b3b5-233e6cc2ff0f.bam'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': '62c0c6fe-99f8-4ff7-b3b5-233e6cc2ff0f.bam'}],
'landingPage': 'https://portal.kidsfirstdrc.org/file/GF_000VDK42'}}],
'types': [{'@type': 'DataType',
'information': {'@type': 'Annotation', 'value': 'Aligned Reads'}}],
'extraProperties': [{'@type': 'CategoryValuesPair',
'category': 'tissue',
'values': [{'@type': 'Annotation', 'value': 'Tumor'}]},
{'@type': 'CategoryValuesPair',
'category': 'diagnosis',
'values': [{'@type': 'Annotation',
'value': 'Brainstem glioma- Diffuse intrinsic pontine glioma, Brainstem glioma- Diffuse intrinsic pontine glioma'}]},
{'@type': 'CategoryValuesPair',
'category': 'proband',
'values': [{'@type': 'Annotation', 'value': 'Yes'}]}]},
{'@type': 'Dataset',
'identifier': {'@type': 'Identifier',
'identifier': '16712090-50f7-4cd1-bf2d-90ce989c2139',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'storedIn': {'@type': 'DataRepository', 'name': 'gen3'},
'producedBy': {'@type': 'Study',
'identifier': {'@type': 'Identifier',
'identifier': 'SD_M3DBXD12',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'name': 'Pediatric Brain Tumor Atlas: PNOC'},
'isAbout': [{'@type': 'BiologicalEntity',
'identifier': {'@type': 'Identifier',
'identifier': 'BS_6DT506HY, BS_5DPMQQVG',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'A08692-T.WXS, A08691-N.WXS'},
{'@type': 'AlternateIdentifier', 'identifier': 'A08713, A08710'}]},
{'@type': 'StudyGroup',
'identifier': {'@type': 'Identifier',
'identifier': 'PT_NK8A49X5',
'identifierSource': 'https://portal.kidsfirstdrc.org/participant/'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'P-06'}]}],
'distributions': [{'identifier': {'@type': 'Identifier',
'identifier': 'GF_000WBJCD',
'identifierSource': 'https://portal.kidsfirstdrc.org/file/'},
'@type': 'DatasetDistribution',
'formats': ['maf'],
'size': 122552,
'unit': {'@type': 'Annotation', 'value': 'bytes'},
'access': {'@type': 'Access',
'identifier': {'@type': 'Identifier',
'identifier': '77af1324-3754-4e34-a208-d1342a2f2ca6.mutect2_somatic.vep.maf'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'harmonized/simple-variants/77af1324-3754-4e34-a208-d1342a2f2ca6.mutect2_somatic.vep.maf'}],
'landingPage': 'https://portal.kidsfirstdrc.org/file/GF_000WBJCD'}}],
'types': [{'@type': 'DataType',
'information': {'@type': 'Annotation',
'value': 'Annotated Somatic Mutations'}}],
'extraProperties': [{'@type': 'CategoryValuesPair',
'category': 'tissue',
'values': [{'@type': 'Annotation', 'value': 'Tumor, Normal'}]},
{'@type': 'CategoryValuesPair',
'category': 'diagnosis',
'values': [{'@type': 'Annotation',
'value': 'Brainstem glioma- Diffuse intrinsic pontine glioma'}]},
{'@type': 'CategoryValuesPair',
'category': 'proband',
'values': [{'@type': 'Annotation', 'value': 'Yes'}]}]},
{'@type': 'Dataset',
'identifier': {'@type': 'Identifier',
'identifier': '49f7aead-ce23-4c38-b566-1d99cb5a5435',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'storedIn': {'@type': 'DataRepository', 'name': 'gen3'},
'producedBy': {'@type': 'Study',
'identifier': {'@type': 'Identifier',
'identifier': 'SD_M3DBXD12',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'name': 'Pediatric Brain Tumor Atlas: PNOC'},
'isAbout': [{'@type': 'BiologicalEntity',
'identifier': {'@type': 'Identifier',
'identifier': 'BS_XGDPK33A',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'A19649-T.RNA-Seq'},
{'@type': 'AlternateIdentifier', 'identifier': 'A19683'}]},
{'@type': 'StudyGroup',
'identifier': {'@type': 'Identifier',
'identifier': 'PT_G16VK7FR',
'identifierSource': 'https://portal.kidsfirstdrc.org/participant/'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'P-37'}]}],
'distributions': [{'identifier': {'@type': 'Identifier',
'identifier': 'GF_001JWT9N',
'identifierSource': 'https://portal.kidsfirstdrc.org/file/'},
'@type': 'DatasetDistribution',
'formats': ['pdf'],
'size': 5779507,
'unit': {'@type': 'Annotation', 'value': 'bytes'},
'access': {'@type': 'Access',
'identifier': {'@type': 'Identifier',
'identifier': '9f586dc1-1df8-4f59-9a01-803141fffb94.arriba.fusions.pdf'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'harmonized/gene-fusions/9f586dc1-1df8-4f59-9a01-803141fffb94.arriba.fusions.pdf'}],
'landingPage': 'https://portal.kidsfirstdrc.org/file/GF_001JWT9N'}}],
'types': [{'@type': 'DataType',
'information': {'@type': 'Annotation', 'value': 'Gene Fusions'}}],
'extraProperties': [{'@type': 'CategoryValuesPair',
'category': 'tissue',
'values': [{'@type': 'Annotation', 'value': 'Tumor'}]},
{'@type': 'CategoryValuesPair',
'category': 'diagnosis',
'values': [{'@type': 'Annotation',
'value': 'Brainstem glioma- Diffuse intrinsic pontine glioma'}]},
{'@type': 'CategoryValuesPair',
'category': 'proband',
'values': [{'@type': 'Annotation', 'value': 'Yes'}]}]},
{'@type': 'Dataset',
'identifier': {'@type': 'Identifier',
'identifier': 'c4c9d542-21fb-487d-ac07-916d466774a8',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'storedIn': {'@type': 'DataRepository', 'name': 'gen3'},
'producedBy': {'@type': 'Study',
'identifier': {'@type': 'Identifier',
'identifier': 'SD_46SK55A3',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'name': 'Kids First: Congenital Diaphragmatic Hernia'},
'isAbout': [{'@type': 'BiologicalEntity',
'identifier': {'@type': 'Identifier',
'identifier': 'BS_BKH9S8YN',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'CDH14-0006'},
{'@type': 'AlternateIdentifier', 'identifier': 'CDH14-0006'}]},
{'@type': 'StudyGroup',
'identifier': {'@type': 'Identifier',
'identifier': 'PT_2HN13G42',
'identifierSource': 'https://portal.kidsfirstdrc.org/participant/'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'CDH14-0006'}]}],
'distributions': [{'identifier': {'@type': 'Identifier',
'identifier': 'GF_002DRSGP',
'identifierSource': 'https://portal.kidsfirstdrc.org/file/'},
'@type': 'DatasetDistribution',
'formats': ['cram'],
'size': 23537329440,
'unit': {'@type': 'Annotation', 'value': 'bytes'},
'access': {'@type': 'Access',
'identifier': {'@type': 'Identifier', 'identifier': 'CDH14-0006.cram'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 's3://kf-study-us-east-1-prd-sd-46sk55a3/source/GMKF_Gabriel_Chung_CDH_WGS/RP-1370/WGS/CDH14-0006/v2/CDH14-0006.cram'}],
'landingPage': 'https://portal.kidsfirstdrc.org/file/GF_002DRSGP'}}],
'types': [{'@type': 'DataType',
'information': {'@type': 'Annotation', 'value': 'Aligned Reads'}}],
'extraProperties': [{'@type': 'CategoryValuesPair',
'category': 'tissue',
'values': [{'@type': 'Annotation', 'value': 'Normal'}]},
{'@type': 'CategoryValuesPair',
'category': 'diagnosis',
'values': [{'@type': 'Annotation',
'value': 'congential diaphragmatic hernia'}]},
{'@type': 'CategoryValuesPair',
'category': 'proband',
'values': [{'@type': 'Annotation', 'value': 'Yes'}]}]},
{'@type': 'Dataset',
'identifier': {'@type': 'Identifier',
'identifier': '02642bc8-ae46-45a1-8932-2765cf1df480',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'storedIn': {'@type': 'DataRepository', 'name': 'gen3'},
'producedBy': {'@type': 'Study',
'identifier': {'@type': 'Identifier',
'identifier': 'SD_46SK55A3',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'name': 'Kids First: Congenital Diaphragmatic Hernia'},
'isAbout': [{'@type': 'BiologicalEntity',
'identifier': {'@type': 'Identifier',
'identifier': 'BS_MP5P3ZPH',
'identifierSource': 'https://portal.kidsfirstdrc.org/'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'CDH4-84F'},
{'@type': 'AlternateIdentifier', 'identifier': 'CDH4-84F'}]},
{'@type': 'StudyGroup',
'identifier': {'@type': 'Identifier',
'identifier': 'PT_MH56TZJD',
'identifierSource': 'https://portal.kidsfirstdrc.org/participant/'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 'CDH4-84F'}]}],
'distributions': [{'identifier': {'@type': 'Identifier',
'identifier': 'GF_004J173A',
'identifierSource': 'https://portal.kidsfirstdrc.org/file/'},
'@type': 'DatasetDistribution',
'formats': ['cram'],
'size': 17290282717,
'unit': {'@type': 'Annotation', 'value': 'bytes'},
'access': {'@type': 'Access',
'identifier': {'@type': 'Identifier', 'identifier': 'CDH4-84F.cram'},
'alternateIdentifiers': [{'@type': 'AlternateIdentifier',
'identifier': 's3://kf-study-us-east-1-prd-sd-46sk55a3/source/GMKF_Gabriel_Chung_CDH_WGS/RP-1370/WGS/CDH4-84F/v2/CDH4-84F.cram'}],
'landingPage': 'https://portal.kidsfirstdrc.org/file/GF_004J173A'}}],
'types': [{'@type': 'DataType',
'information': {'@type': 'Annotation', 'value': 'Aligned Reads'}}],
'extraProperties': [{'@type': 'CategoryValuesPair',
'category': 'tissue',
'values': [{'@type': 'Annotation', 'value': 'Normal'}]},
{'@type': 'CategoryValuesPair',
'category': 'proband',
'values': [{'@type': 'Annotation', 'value': 'No'}]}]}]}
Now we see, with some mapping effort, we were able to get all of the metadata
from the file manifest table into DATS. It is important to note that there are
manys fields missing including license, authorship information, and more. These fields
need to be found from other places to further complete and improve this model.
With our newly created object, let’s check to make sure we did not make any mistakes!
For this purpose, just as we can use json-schema for auto completion help in our editor,
we can also use it for programatic validation of our dats object.
from jsonschema import Draft4Validator
# Get the first record
record = dats['@graph'][0]
# Validate it against DATS dataset schema
validator = Draft4Validator({'$ref': 'https://raw.githubusercontent.com/datatagsuite/schema/master/dataset_schema.json'})
for error in validator.iter_errors(record):
display(error.message)
"{'@type': 'BiologicalEntity', 'identifier': {'@type': 'Identifier', 'identifier': 'BS_A7Q8G0Y1', 'identifierSource': 'https://portal.kidsfirstdrc.org/'}, 'alternateIdentifiers': [{'@type': 'AlternateIdentifier', 'identifier': '7316-126-T-112502.RNA-Seq'}, {'@type': 'AlternateIdentifier', 'identifier': '746063'}]} is not valid under any of the given schemas"
"{'@type': 'StudyGroup', 'identifier': {'@type': 'Identifier', 'identifier': 'PT_PR4YBBH3', 'identifierSource': 'https://portal.kidsfirstdrc.org/participant/'}, 'alternateIdentifiers': [{'@type': 'AlternateIdentifier', 'identifier': 'C29274'}]} is not valid under any of the given schemas"
"'title' is a required property"
"'creators' is a required property"
Uh-oh; we’ve got some errors.
Let’s fix them and try again.
For readability, the changes we had to make below are here:
@@ -1,6 +1,7 @@
def dats_from_record(record):
return {
'@type': 'Dataset',
+ 'title': record['Study Name'],
'identifier': {
'@type': 'Identifier',
'identifier': record['Latest DID'],
@@ -10,6 +11,12 @@
'@type': 'DataRepository',
'name': record['Repository'],
},
+ 'creators': [
+ {
+ "@type": "Organization",
+ "name": "KidsFirst",
+ }
+ ],
'producedBy': {
'@type': 'Study',
'identifier': {
@@ -22,6 +29,8 @@
'isAbout': [
{
'@type': 'BiologicalEntity',
+ # NOTE: name is a required field
+ 'name': record['Biospecimen ID'],
'identifier': {
'@type': 'Identifier',
'identifier': record['Biospecimen ID'],
@@ -42,6 +51,8 @@
},
{
'@type': 'StudyGroup',
+ # NOTE: name is a required field
+ 'name': record['Participants ID'],
'identifier': {
# NOTE: Ideally, `${identifierSource}${identifier}` resolves to a landing page for this entity
'@type': 'Identifier',
@@ -69,7 +80,7 @@
'formats': [
record['File Format'],
],
- 'size': record['File Size'],
+ 'size': int(record['File Size']),
'unit': {
'@type': 'Annotation',
'value': 'bytes',
@@ -141,4 +152,4 @@
],
} if record['Proband'] != '--' else None, # Don't create entries for junk,
])],
- }
+ }
You can see that we put in an invalid type and were missing some fields. In some cases, we need to add metadata that wasn’t in the original table such as metadata about our own organization!
This is relevant in a catalog of many datasets but often isn’t present in your own data; it’s best if you determine how your own data will link back to your organization, than us trying to figure it out! That’s why DATS requires that metadata.
def dats_from_record(record):
return {
'@type': 'Dataset',
'title': record['Study Name'],
'identifier': {
'@type': 'Identifier',
'identifier': record['Latest DID'],
'identifierSource': 'https://portal.kidsfirstdrc.org/',
},
'storedIn': {
'@type': 'DataRepository',
'name': record['Repository'],
},
'creators': [
{
"@type": "Organization",
"name": "KidsFirst",
}
],
'producedBy': {
'@type': 'Study',
'identifier': {
'@type': 'Identifier',
'identifier': record['Study ID'],
'identifierSource': 'https://portal.kidsfirstdrc.org/',
},
'name': record['Study Name'],
},
'isAbout': [
{
'@type': 'BiologicalEntity',
# NOTE: name is a required field
'name': record['Biospecimen ID'],
'identifier': {
'@type': 'Identifier',
'identifier': record['Biospecimen ID'],
'identifierSource': 'https://portal.kidsfirstdrc.org/',
},
'alternateIdentifiers': [
{
'@type': 'AlternateIdentifier',
'identifier': record['Sample External ID'],
# NOTE: Preferred identifierSource with globally unique semantic URI
},
{
'@type': 'AlternateIdentifier',
'identifier': record['Aliquot External ID'],
# NOTE: Preferred identifierSource with globally unique semantic URI
},
],
},
{
'@type': 'StudyGroup',
# NOTE: name is a required field
'name': record['Participants ID'],
'identifier': {
# NOTE: Ideally, `${identifierSource}${identifier}` resolves to a landing page for this entity
'@type': 'Identifier',
'identifier': record['Participants ID'],
'identifierSource': 'https://portal.kidsfirstdrc.org/participant/',
},
'alternateIdentifiers': [
{
'@type': 'AlternateIdentifier',
'identifier': record['Participant External ID'],
# NOTE: Preferred identifierSource with globally unique semantic URI
},
],
},
],
'distributions': [
{
'identifier': {
# NOTE: Ideally, `${identifierSource}${identifier}` resolves to a landing page for this entity
'@type': 'Identifier',
'identifier': record['File ID'],
'identifierSource': 'https://portal.kidsfirstdrc.org/file/',
},
'@type': 'DatasetDistribution',
'formats': [
record['File Format'],
],
'size': int(record['File Size']),
'unit': {
'@type': 'Annotation',
'value': 'bytes',
# NOTE: Preferred valueIRI with globally unique semantic URI
},
'access': {
'@type': 'Access',
'identifier': {
'@type': 'Identifier',
'identifier': record['File Name'],
},
'alternateIdentifiers': [
{
'@type': 'AlternateIdentifier',
'identifier': record['File External ID'],
# NOTE: Preferred identifierSource with globally unique semantic URI
},
],
'landingPage': 'https://portal.kidsfirstdrc.org/file/' + record['File ID'],
# NOTE: Ideally accessURL would be specified
}
}
],
'types': [
{
'@type': 'DataType',
'information': {
'@type': 'Annotation',
'value': record['Data Type'],
},
},
],
'extraProperties': [*filter(None, [
# Metadata that doesn't fit anywhere else in DATS but may be relevant
{
'@type': 'CategoryValuesPair',
'category': 'tissue',
# NOTE: Preferred categoryIRI with globally unique semantic URI
'values': [
{
'@type': 'Annotation',
'value': record['Tissue Type (Source Text)'],
# NOTE: Preferred valueIRI with globally unique semantic URI
}
]
} if record['Tissue Type (Source Text)'] != '--' else None,
{
'@type': 'CategoryValuesPair',
'category': 'diagnosis',
# NOTE: Preferred categoryIRI with globally unique semantic URI
'values': [
{
'@type': 'Annotation',
'value': record['Diagnosis (Source Text)'],
# NOTE: Preferred valueIRI with globally unique semantic URI
}
]
} if record['Diagnosis (Source Text)'] != '--' else None, # Don't create entries for junk
{
'@type': 'CategoryValuesPair',
'category': 'proband',
# NOTE: Preferred categoryIRI with globally unique semantic URI
'values': [
{
'@type': 'Annotation',
'value': record['Proband'],
# NOTE: Preferred valueIRI with globally unique semantic URI
},
],
} if record['Proband'] != '--' else None, # Don't create entries for junk,
])],
}
# Converting each element to DATS
dats = {
# schema.org context, gives RDF meaning to `@type` and `predicates` as defined by schema.org
'@context': 'http://w3id.org/dats/context/sdo/dataset_sdo_context.jsonld',
'@graph': [
dats_from_record(record)
for _, record in df.head().iterrows()
]
}
# Let's validate *all records*
record = dats['@graph'][0]
# Validate it against DATS dataset schema
validator = Draft4Validator({
'$ref': 'https://raw.githubusercontent.com/datatagsuite/schema/master/dataset_schema.json'
})
for record in dats['@graph']:
for error in validator.iter_errors(record):
display({ 'title': record['title'], 'error': error.message })
Conclusion¶
As hoped, everything validates and we have successfully produced DATS.
Though we now know our DATS is “valid”, we’re still not done. As with everything
there are levels; the more fields we fill out in the DATS the better off
we will be. This is where a FAIR assessment comes in – we can write metrics
that also speak DATS, but are looking for presence of certain fields,
or checking that our identifier can actually be verified against the given identifierSource
metadata attributes.
Nonetheless, we have taken a step in the right direction. Future recipes will discuss performing FAIR assessments on this DATS, converting it to CFDE’s C2M2 Frictionless Metadata model and more!
